 One of the main themes of my research is understanding how different evolutionary processes and contexts influence genome divergence during speciation. I take a comparative approach to address questions in evolution by sampling replicate population pairs with varying levels of divergence. These comparisons provide multiple snapshots across the continuum building a more complete picture of the speciation process. Additionally, I combine modern genomic tools with phenotypic trait data and spatial analyses to complement the comparative perspective with an integrative approach. The main goal is to broaden our understanding of the dynamic process of speciation by addressing long-standing questions and effecting novel pursuits in the field of evolutionary biology.
One of the main themes of my research is understanding how different evolutionary processes and contexts influence genome divergence during speciation. I take a comparative approach to address questions in evolution by sampling replicate population pairs with varying levels of divergence. These comparisons provide multiple snapshots across the continuum building a more complete picture of the speciation process. Additionally, I combine modern genomic tools with phenotypic trait data and spatial analyses to complement the comparative perspective with an integrative approach. The main goal is to broaden our understanding of the dynamic process of speciation by addressing long-standing questions and effecting novel pursuits in the field of evolutionary biology.

University of California, Berkeley
Museum of Vertebrate Zoology
California, USA
Undergraduate
My first research experience focused on phylogeography and phylogenetics of various African bird taxa under the supervision of Rauri Bowie.

Australian National University
Canberra, Australia
PhD student
I completed my PhD in 2017 supervised by Craig Moritz at ANU and Leo Joseph at CSIRO. My thesis is entitled “Speciation genomics in the Australian Meliphagoidea”.

Ludwig-Maximillians-Universität München
Munich, Germany
Postdoctoral researcher
I worked as a postdoc in the lab of Jochen Wolf to develop methods to estimate recombination rate variation across the genome.

Museum für Naturkunde
Berlin, Germany
Research scientist
Currently, I am a research scientist specializing in museomics and junior group leader. The focus of my research group is speciation, population genetics, and comparative biogeography.
Research
SPECIATION GENOMICS
Genome divergence between populations is governed by selection, drift, and gene flow. During speciation, these mechanisms are influenced by a suite of processes including demographic history, geography, interpopulation interactions, and incompatibilities. My research aims to shed light on the influence of these processes during speciation using a comparative approach sampling broadly at different stages of the speciation continuum.

COMPARATIVE GENOMICS
Combining short-read and long-read technologies have allowed us to assemble a high quality genome for the superb fairywren (Malurus cyaneus). Not only does this genome compliment a long-term ecological study of this species but also provide a resource for comparison with existing genome assemblies. I aim to take advantage of these resources to further understand genome evolution both in a micro- and macroevolutionary scale.
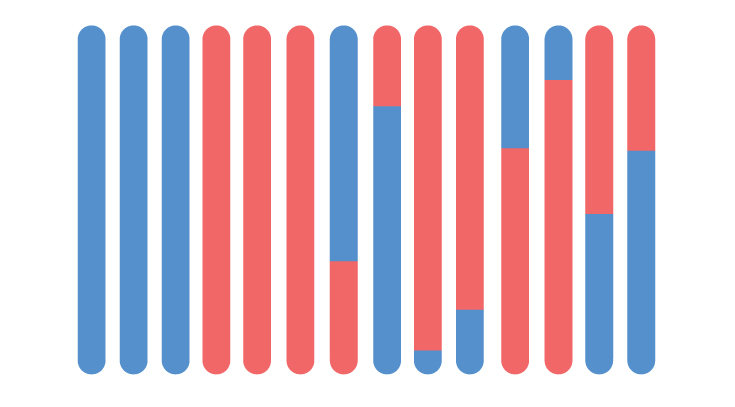
HYBRID & SUTURE ZONES
Hybrid zones are said provide a “natural experiment to the evolutionary process”. These zones allow researchers to observe interpopulation interactions and estimate the geographic extent of gene flow between diverging populations. For my research, I use codistributed contact zones (“suture zones”) between populations of varying divergence levels. These multiple replicate experiments allow us to infer how divergence and gene flow change through both space and time in a shared geographic context.

GENOME EVOLUTION
Whole genome sequencing has revealed the variation of divergence across the genome. This variation is hypothesized to result from variation of drift, gene flow, and selection within the genomes interacting with genome architecture. I am interested in how genomic features such as recombination rate variation or structural variation can influence interact with these forces, in turn influencing divergence during early and late stages of speciation.